Bacterial RNAs
From Wiki CEINGE
| Revision as of 13:45, 18 June 2007 (edit) Luca (Talk | contribs) ← Previous diff |
Current revision (13:15, 22 June 2007) (edit) (undo) Leandra (Talk | contribs) |
||
| (7 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
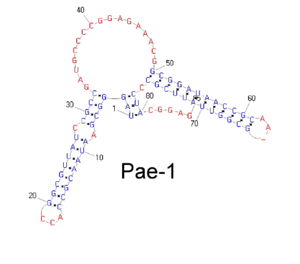
| - | + | [[Image:Bacterial_pae.jpg|300px|right|thumb|PAE-1 bacterial family secondary structure]] | |
| - | + | Bacterial genomes are generally compact and most of their sequence is involved in protein coding, but a growing number of sequences, mostly located within the intergenic regions, have been shown to play a role in the control of gene expression. Many of these sequences are active as RNA and often contain simple stem-loop structures (SLS), essential to their functionality. SLSs have been found also in repetitive sequences in several bacterial genomes, even if only in few cases a clear biological function was assessed. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Starting from this considerations we decided to perform a systematic analysis of these elements and to identify | ||
| + | all the repeated sequence families able to share a common SLS. The project started with the identification of all the sequences able to fold in a SLS manner from a set of 40 wholly-sequenced genomes representative of the bacterial world. SLSs were extracted, annotated and stored in a relational database. A first result of this project was to demonstrate that SLSs found in natural genomes are constantly more numerous and stable than those expected to randomly form in sequences of comparable size and base composition. It is therefore possible that there is a selective pressure in some microorganisms to preserve these sequences because of their biological functions. | ||
| + | A second analysis based on clustering procedures revealed that most of analyzed genomes have SLSs that can be grouped by sequence similarity. Such SLSs, which correspond to a very little fraction of the starting population, have a substantially higher aptitude to fold into a stable secondary structure than the initial set. | ||
| + | This procedure allowed to identify a large collection of families of repeated stem-loop containing sequences. Secondary structure analysis revealed for many of them the presence of a conserved secondary structure, possibly linked to their biological function. | ||
| Line 14: | Line 11: | ||
| * [http://www.biomedcentral.com/1471-2164/7/170 PETRILLO M., SILVESTRO G., DI NOCERA PP., BOCCIA A. and PAOLELLA G. Stem-loop structures in prokaryotic genomes (2006) BMC GENOMICS 2006, 7:170] | * [http://www.biomedcentral.com/1471-2164/7/170 PETRILLO M., SILVESTRO G., DI NOCERA PP., BOCCIA A. and PAOLELLA G. Stem-loop structures in prokaryotic genomes (2006) BMC GENOMICS 2006, 7:170] | ||
| * COZZUTO L., PETRILLO M., SILVESTRO G., DI NOCERA PP. and PAOLELLA G. Systematic identification of stem-loop containing sequence families in bacterial genomes SUBMITTED. | * COZZUTO L., PETRILLO M., SILVESTRO G., DI NOCERA PP. and PAOLELLA G. Systematic identification of stem-loop containing sequence families in bacterial genomes SUBMITTED. | ||
| + | |||
| + | |||
| + | {{footer reslines}} | ||
Current revision
Bacterial genomes are generally compact and most of their sequence is involved in protein coding, but a growing number of sequences, mostly located within the intergenic regions, have been shown to play a role in the control of gene expression. Many of these sequences are active as RNA and often contain simple stem-loop structures (SLS), essential to their functionality. SLSs have been found also in repetitive sequences in several bacterial genomes, even if only in few cases a clear biological function was assessed.
Starting from this considerations we decided to perform a systematic analysis of these elements and to identify all the repeated sequence families able to share a common SLS. The project started with the identification of all the sequences able to fold in a SLS manner from a set of 40 wholly-sequenced genomes representative of the bacterial world. SLSs were extracted, annotated and stored in a relational database. A first result of this project was to demonstrate that SLSs found in natural genomes are constantly more numerous and stable than those expected to randomly form in sequences of comparable size and base composition. It is therefore possible that there is a selective pressure in some microorganisms to preserve these sequences because of their biological functions. A second analysis based on clustering procedures revealed that most of analyzed genomes have SLSs that can be grouped by sequence similarity. Such SLSs, which correspond to a very little fraction of the starting population, have a substantially higher aptitude to fold into a stable secondary structure than the initial set. This procedure allowed to identify a large collection of families of repeated stem-loop containing sequences. Secondary structure analysis revealed for many of them the presence of a conserved secondary structure, possibly linked to their biological function.
[edit] References
- PETRILLO M., SILVESTRO G., DI NOCERA PP., BOCCIA A. and PAOLELLA G. Stem-loop structures in prokaryotic genomes (2006) BMC GENOMICS 2006, 7:170
- COZZUTO L., PETRILLO M., SILVESTRO G., DI NOCERA PP. and PAOLELLA G. Systematic identification of stem-loop containing sequence families in bacterial genomes SUBMITTED.