CAPRI
From Wiki CEINGE
| Revision as of 13:10, 15 June 2007 (edit) Luca (Talk | contribs) ← Previous diff |
Revision as of 13:34, 15 June 2007 (edit) (undo) Luca (Talk | contribs) Next diff → |
||
| Line 11: | Line 11: | ||
| * a lateral menu bar (2), which allows to: open other pages, load data from disk, servers or databases, copy the visible data to a new page and select that sequences that have to be displayed. | * a lateral menu bar (2), which allows to: open other pages, load data from disk, servers or databases, copy the visible data to a new page and select that sequences that have to be displayed. | ||
| * a top menu bar (3), which allows to access the installed analysis tool and which can dynamically changes depending on the number of data. | * a top menu bar (3), which allows to access the installed analysis tool and which can dynamically changes depending on the number of data. | ||
| - | + | Clicking on a tool link in a menu, a new window appears showing all the modifiable options of such program and on the top-right a link to its documentation. | |
| Currently CAPRI implements five kinds of page to analyze five different biological data: DNA, protein, alignment, Hidden Markov Model (HMM) and phylogenetic tree; each page contains links to access to all the programs available for the analysis of respective input data. | Currently CAPRI implements five kinds of page to analyze five different biological data: DNA, protein, alignment, Hidden Markov Model (HMM) and phylogenetic tree; each page contains links to access to all the programs available for the analysis of respective input data. | ||
| - | The result of each analysis is displayed in a new page, which type is in agreement with the output data: for example, DNA sequences are aligned within the 'DNA page' and the resulting alignment is provided in an 'alignment page'. | + | The result of each analysis is displayed in a new page, which type is in agreement with the output data: for example, DNA sequences are aligned within the 'DNA page' and the resulting alignment is provided in an 'alignment page'. Some tool provides more than one output. In this case the output page has a further button on the top-right corner which, once clicked, allows to switch to the other data. |
| === DNA Page === | === DNA Page === | ||
| Line 21: | Line 21: | ||
| :''Analyze'': computing of GC content and temperature melting | :''Analyze'': computing of GC content and temperature melting | ||
| :''Process'': making the complement and the translation in protein | :''Process'': making the complement and the translation in protein | ||
| - | :''Search'': finding restriction enzyme (RE) sites, patterns, open reading frames (ORFs), PCR primers | + | :''Search'': finding restriction enzyme (RE) sites, patterns, open reading frames (ORFs), PCR primers and siRNAs |
| - | :''Extract'': extracting a sequence fragment with the given coordinates | + | :''Extract'': extracting a sequence fragment with the given coordinates |
| - | :''SearchDB'': searching in sequence databases | + | :''SearchDB'': searching in sequence databases. |
| More used functions in multi sequence analysis : | More used functions in multi sequence analysis : | ||
| :''Draw'': drawing a dot-plot, which shows sequence similarity between sequences | :''Draw'': drawing a dot-plot, which shows sequence similarity between sequences | ||
| Line 31: | Line 31: | ||
| Protein Page allows the analysis of proteic sequences. Top menu bar behaviour is similar to the DNA page. | Protein Page allows the analysis of proteic sequences. Top menu bar behaviour is similar to the DNA page. | ||
| More used functions in single sequence analysis divided for menu: | More used functions in single sequence analysis divided for menu: | ||
| - | :''Analyze'': | + | :''Analyze'': analysis of proteolytic enzyme or reagent cleavage digest, of charge, isoelectric point and secondary structure |
| - | :''Process'': making the | + | :''Process'': making the back-translation and the shuffling |
| - | :''Search'': | + | :''Search'': searching pattern, coiled coil, transmembrane tract, cleavage and antigenic sites. |
| - | :''Extract'': extracting a sequence fragment with the given coordinates | + | :''Extract'': extracting a sequence fragment with the given coordinates |
| - | :''SearchDB | + | :''SearchDB: searching in sequence databases. |
| More used functions in multi sequence analysis : | More used functions in multi sequence analysis : | ||
| :''Draw'': drawing a dot-plot, which shows sequence similarity between sequences | :''Draw'': drawing a dot-plot, which shows sequence similarity between sequences | ||
Revision as of 13:34, 15 June 2007
CAPRI (Common Application Program Remote Interface) is a web interface for accessing all the major tools currently installed at CEINGE. It was developed at CEINGE bionformatics lab and can be used only by registered users. CAPRI project was born to guarantee the researchers an easy access to many widely used sequence analysis tools, such as BLAST, FastA or even packages like EMBOSS, which were originally developed on a command line interface. CAPRI resembles a typical local desktop application where the user selects the sequences and uses a number of menus to access the various functions but, in reality, it takes advantage of the processing power and the databases available on the CEINGE's remote servers. Unlike other similar tools CAPRI does not require to install any software. Click here to access to CAPRI.
Contents |
Tool description
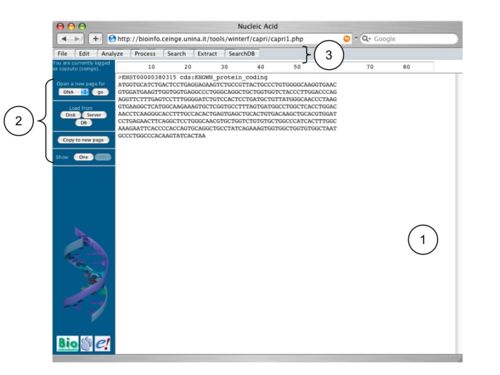
A page of CAPRI is a web-page consisting of:
- a text-area (1), in which user can insert and edit his data. For example, it is possible to paste directly one or more sequences in this area.
- a lateral menu bar (2), which allows to: open other pages, load data from disk, servers or databases, copy the visible data to a new page and select that sequences that have to be displayed.
- a top menu bar (3), which allows to access the installed analysis tool and which can dynamically changes depending on the number of data.
Clicking on a tool link in a menu, a new window appears showing all the modifiable options of such program and on the top-right a link to its documentation. Currently CAPRI implements five kinds of page to analyze five different biological data: DNA, protein, alignment, Hidden Markov Model (HMM) and phylogenetic tree; each page contains links to access to all the programs available for the analysis of respective input data. The result of each analysis is displayed in a new page, which type is in agreement with the output data: for example, DNA sequences are aligned within the 'DNA page' and the resulting alignment is provided in an 'alignment page'. Some tool provides more than one output. In this case the output page has a further button on the top-right corner which, once clicked, allows to switch to the other data.
DNA Page
DNA page is the CAPRI default page that allows to analyze nucleic acid sequences like DNA or RNA. Top menu bar changes if one, two or more sequences are inserted and analyzed at the same time, indicating which tools are available for the analysis.
More used functions in single sequence analysis divided for menu:
- Analyze: computing of GC content and temperature melting
- Process: making the complement and the translation in protein
- Search: finding restriction enzyme (RE) sites, patterns, open reading frames (ORFs), PCR primers and siRNAs
- Extract: extracting a sequence fragment with the given coordinates
- SearchDB: searching in sequence databases.
More used functions in multi sequence analysis :
- Draw: drawing a dot-plot, which shows sequence similarity between sequences
- Search: searching matches by profile
- Align: aligning sequences with the most popular tools (ClustalW, matcher, strecher).
Protein Page
Protein Page allows the analysis of proteic sequences. Top menu bar behaviour is similar to the DNA page. More used functions in single sequence analysis divided for menu:
- Analyze: analysis of proteolytic enzyme or reagent cleavage digest, of charge, isoelectric point and secondary structure
- Process: making the back-translation and the shuffling
- Search: searching pattern, coiled coil, transmembrane tract, cleavage and antigenic sites.
- Extract: extracting a sequence fragment with the given coordinates
- SearchDB: searching in sequence databases.
More used functions in multi sequence analysis :
- Draw: drawing a dot-plot, which shows sequence similarity between sequences
- Search: searching matches by profile
- Align: aligning sequences with the most popular tools (ClustalW, matcher, strecher).
Alignment Page
With this page it is possible to analyze sequence alignments.
HMM Page
This page allows the analysis of Hidden Markov Models. Profile HMMs turn a multiple sequence alignment into a position-specific scoring system suitable for searching databases for remotely homologous sequences. Profile HMM analyses complement standard pairwise comparison methods for large-scale sequence analysis.
TREE Page
This page allow the analysis of phylogenetic tree.