DG-CST
From Wiki CEINGE
| Revision as of 16:02, 20 June 2007 (edit) Mauro (Talk | contribs) ← Previous diff |
Revision as of 16:02, 20 June 2007 (edit) (undo) Mauro (Talk | contribs) Next diff → |
||
| Line 5: | Line 5: | ||
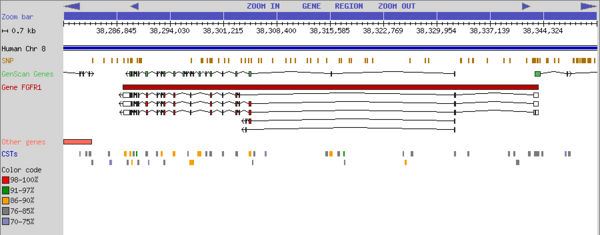
| The database is available at http://dgcst.ceinge.unina.it, where it is possible to query and select CSTs respect to their sequence and annotations. A graphic browser allows direct visualization of the CSTs and related annotations within the context of the relative gene and its transcripts. | The database is available at http://dgcst.ceinge.unina.it, where it is possible to query and select CSTs respect to their sequence and annotations. A graphic browser allows direct visualization of the CSTs and related annotations within the context of the relative gene and its transcripts. | ||
| - | [[Image:Dgcst.jpg|center|DG-CST Graphic browser]] | + | [[Image:Dgcst.jpg|center|600px|DG-CST Graphic browser]] |
| ===Available annotations of CSTs=== | ===Available annotations of CSTs=== | ||
Revision as of 16:02, 20 June 2007
The DG-CST (Disease Gene Conserved Sequence Tags) database is a collection of conserved sequence elements, identified by a systematic genomic sequence comparison between a set of human more than 1000 genes involved in the pathogenesis of genetic disorders and their murine counterparts.
How to reach and use DG-CST
The database is available at http://dgcst.ceinge.unina.it, where it is possible to query and select CSTs respect to their sequence and annotations. A graphic browser allows direct visualization of the CSTs and related annotations within the context of the relative gene and its transcripts.
Available annotations of CSTs
Human and Mouse genomic sequences were compared by BLASTZ, an independent implementation of the Gapped BLAST algorithm, specifically designed for aligning two long genomic sequences. Sequences longer than 100 and with identity better than 70% were selected as CSTs and imported into the DB. CST counterparts in other species were identified by using BLAST to scan genomes from other species, and selecting on the basis of homology and colinearity. In the database, CSTs are collected together with a large number of annotations, including:
- genomic location, i.e. chromosome, position, relationship with the closest gene and with the selected disease gene (often coincident);
- sequence content, i.e. sequence, length, GC percentage;
- identity between human and mouse sequences, number of gaps, polarity;
- BLAST matches with other CSTs, as well as with other human genomic sequences;
- BLAST matches versus non-redundant nucleotide databases;
- conservation in other species, as assessed by BLAST analysis versus the drafts of fugu chicken, rat and zebrafish genome sequences;
- classification of CSTs in ‘intronic’, ‘intergenic’, ‘exonic’ based on Ensembl gene annotations;
- presence of single nucleotide polymorphisms (SNPs), as reported in Ensembl;
- presence of palindromes, tandem repeats, putative RNA
- secondary structures prediciton;
- presence of putative transcription factor (TF) binding sites;
- presence of palindromes and tandem repeats
In order to identify CSTs potentially representing novel transcribed/coding elements, different tests were performed, such as:
- determination of maximum ORF size;
- presence of putative splice sites, exonic splicing enhancers;
- exon predictions based on GENSCAN;
- BLAST matches with expressed sequence tags (ESTs) and non-redundant protein databases;
References
BLASTZ is available at http://bio.cse.psu.edu.