Image processing
From Wiki CEINGE
| Revision as of 14:07, 20 June 2007 (edit) Concita (Talk | contribs) ← Previous diff |
Revision as of 16:14, 20 June 2007 (edit) (undo) Leandra (Talk | contribs) Next diff → |
||
| Line 1: | Line 1: | ||
| - | The study of the cell motility takes advantage both of morphological investigations of fixed samples and functional | + | Cell motility is a complex phenomenon, which may be explored in a variety of experimental situations, ranging from in vitro cultured cells to migration in whole embryos or in the adult animal. The study of the cell motility takes advantage both of morphological investigations of fixed samples, and of functional analysis of live cells using experimental conditions as soon as possible close to physiological ones. Timelapse-microscopy, in combination with fast optical sectioning techniques, allows to observe and analyze the movement in three dimensions of whole live cells, but also of their subcellular components and of specific molecules at subsequent time steps. By applying video time lapse techniques to the study of live fibroblasts, the role of specific molecules such as oncogenes and components of the extracellular matrix, has been evaluated. |
| - | + | The large number of images and correlated experimental data and results acquired in the time, required to integrate storage and management with the availability of each set of data for further analysis. | |
| + | In order to increase the visibility of some cell structures blurred by redundant fluorescence signal, it's often necessary to apply restoration filters. Deconvolution tool was developed to restore fluorescent acquired images. | ||
| + | To execute statistical analysis of cell motility................ | ||
| + | A web based system able to visualize, analyse and process multidimensional images was also been developed; this tool, named IPROC will integrate all the components previously described, within one global web system. | ||
| - | ''' | + | '''Cell motility tools''' A set of software tools for visualization and statistical evaluation of cellular movements from series of subsequent frames acquired manually or from automated video time-lapse microscopy, has been developed. These tools ([[Cell Motility]]) have been adapted and used to study the sub-cellular structures migration such as endocytosis vesicles, within the cytoplasm. |
| + | The first result contribute to associate to different cell lines specific motion features; but a punctual analysis is also developed to study sub-cellular elements in order to make visible their different ability to move into the cytoplasm in response to distinct stimuli. The tools execute statistical analysis to measure the speed, the direction and the tortuosity of moving elements. | ||
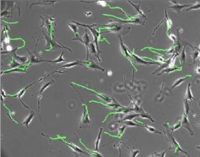
| - | [[Image: | + | [[Image:paths.jpg|center|200px|overlay image and paths]] |
| - | '''Cell motility tools.''' A set of software tools for visualization and statistical evaluation of cellular movements from series of subsequent frames acquired manually or from automated video time-lapse microscopy, has been developed. These tools ([[Cell Motility]]) have been adapted and used to study the sub-cellular structures migration such as endocytosis vesicles, within the cytoplasm. | ||
| - | The first result contribute to associate to different cell lines specific motion features; but a punctual analysis is also developed to study sub-cellular elements in order to make visible their different ability to move into the cytoplasm in response to distinct stimuli. The tools execute statistical analysis to measure the speed, the direction and the tortuosity of moving elements. | ||
| - | [[Image: | + | '''Data bank of biological images and correlated data''' To store and manage the large amount of digital images produced has been developed a relational databank ([[ImagesDB]]) able to connect to each acquisition the correlated experimental data such as cell line, colture conditions and staining methods. The data bank has also a user-friendly web-interface to visualize and update the image data. |
| + | |||
| + | |||
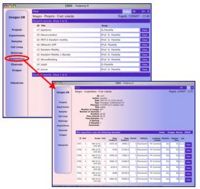
| + | [[Image:ImagesDB.jpg|center|200px|images data bank]] | ||
| + | |||
Revision as of 16:14, 20 June 2007
Cell motility is a complex phenomenon, which may be explored in a variety of experimental situations, ranging from in vitro cultured cells to migration in whole embryos or in the adult animal. The study of the cell motility takes advantage both of morphological investigations of fixed samples, and of functional analysis of live cells using experimental conditions as soon as possible close to physiological ones. Timelapse-microscopy, in combination with fast optical sectioning techniques, allows to observe and analyze the movement in three dimensions of whole live cells, but also of their subcellular components and of specific molecules at subsequent time steps. By applying video time lapse techniques to the study of live fibroblasts, the role of specific molecules such as oncogenes and components of the extracellular matrix, has been evaluated. The large number of images and correlated experimental data and results acquired in the time, required to integrate storage and management with the availability of each set of data for further analysis. In order to increase the visibility of some cell structures blurred by redundant fluorescence signal, it's often necessary to apply restoration filters. Deconvolution tool was developed to restore fluorescent acquired images. To execute statistical analysis of cell motility................ A web based system able to visualize, analyse and process multidimensional images was also been developed; this tool, named IPROC will integrate all the components previously described, within one global web system.
Cell motility tools A set of software tools for visualization and statistical evaluation of cellular movements from series of subsequent frames acquired manually or from automated video time-lapse microscopy, has been developed. These tools (Cell Motility) have been adapted and used to study the sub-cellular structures migration such as endocytosis vesicles, within the cytoplasm.
The first result contribute to associate to different cell lines specific motion features; but a punctual analysis is also developed to study sub-cellular elements in order to make visible their different ability to move into the cytoplasm in response to distinct stimuli. The tools execute statistical analysis to measure the speed, the direction and the tortuosity of moving elements.
Data bank of biological images and correlated data To store and manage the large amount of digital images produced has been developed a relational databank (ImagesDB) able to connect to each acquisition the correlated experimental data such as cell line, colture conditions and staining methods. The data bank has also a user-friendly web-interface to visualize and update the image data.
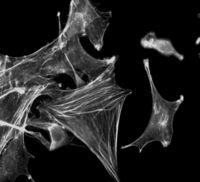
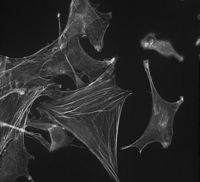
Software to restore digital images. Using optical sectioning technique, a stack of 2D images can be obtained. However, due to the nature of the optical system and non-optimal experimental conditions, acquired raw data are usually sensitive to some distorsions. Raw data have to be restored in order to carry out biological analysis and enhance the visibility of cellular structure otherwise hidden. The images, obtained by a conventional fluorescence microscope, contain light from the all 3D sample-object. this means an evident blurring of the focal plane by “out –of - focus” fluorescence. The reduction of this effect is carried out by a computational deconvolution process. We developed a custom tool to execute the deconvolution algorithm, it attempt to reassign blurred light to its location.
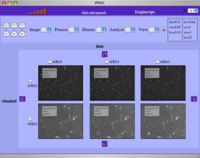
IPROC a web-system to visualize and process biological images A suitable visualization and some opportune image processes may highlight various aspects of the image data. This consideration led to the development of IPROC a web based image visualization and processing system (IPROC). Compatible images range from simple two-dimensional frames, to complex, multidimensional datasets typical of microscopic observation of biological images, springing from variable combinations of time series, z-axis sections, channels recorded at different wavelength and optical arrangement with, optionally, different samples and positions. The program include scrolling, rotation and selection mechanisms to make a variable visualization of a multidimensional image. Processing is obtained by integrating a large library of different unix filters installed on the server, while interactivity is provided by the ability to quickly react to user input via small data requests.
old
Cell motility is a complex phenomenon, which may be explored in a variety of experimental situations, ranging from in vitro cultured cells to cell migration in whole embryos or in the adult animal. By applying video time lapse techniques to the study of live fibroblasts, the role of specific molecules such as oncogenes and components of the extracellular matrix has been evaluated. The need for applying image processing techniques to the study, stimulated the development of a number of computational tools, including a system for centralized storage of experimental images, and a web based image processing tool. Statistical analysis of cell movement and study of multidimensional images may be performed within these systems.